SESAME
SESAME is an open-source Python tool designed to make spatial data analysis, visualization, and exploration accessible to all.
Whether you’re a researcher, student, or enthusiast, SESAME helps you unlock insights from geospatial data with just a few lines of code.
What can you do with the SESAME toolbox?
- Conveniently process and analyze both spatial datasets (e.g. GeoTIFFs) and tabular jurisdictional data (e.g. csv files by country) through a unified set of tools.
- Generate standardized netcdf files from a wide range of spatial input types (e.g. lines, points, polygons)
- Create publication-ready maps and plots.
- Explore spatial and temporal patterns among hundreds of variables in the Human-Earth Atlas.
Getting Started with the Human-Earth Atlas:
- Install SESAME*
- Download the Human-Earth Atlas (Figshare Link)
- Load your spatial data (e.g., land cover, population, climate)
- Use SESAME’s plotting tools to visualize and compare datasets
- Explore spatial and temporal patterns among hundreds of variables in the Human-Earth Atlas.
*Note: SESAME may take up to 2 minutes to load when used for the first time. This will not recur with further use.
Navigating the Atlas:
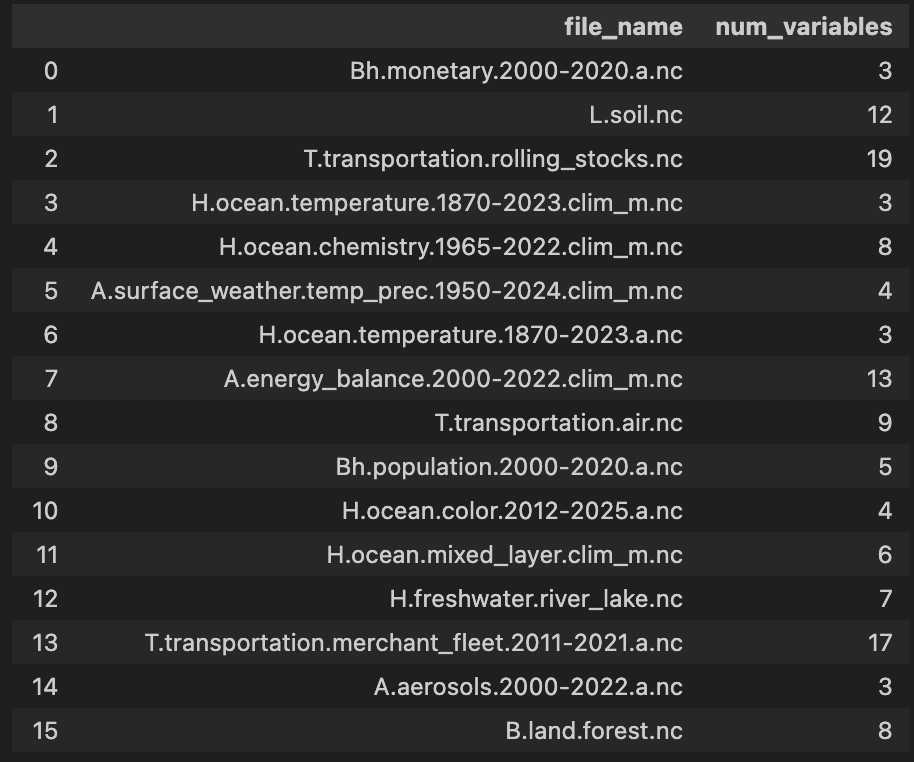
- List the netCDF files in the Human–Earth Atlas
import sesame as ssm
ssm.atlas(directory=atlas)
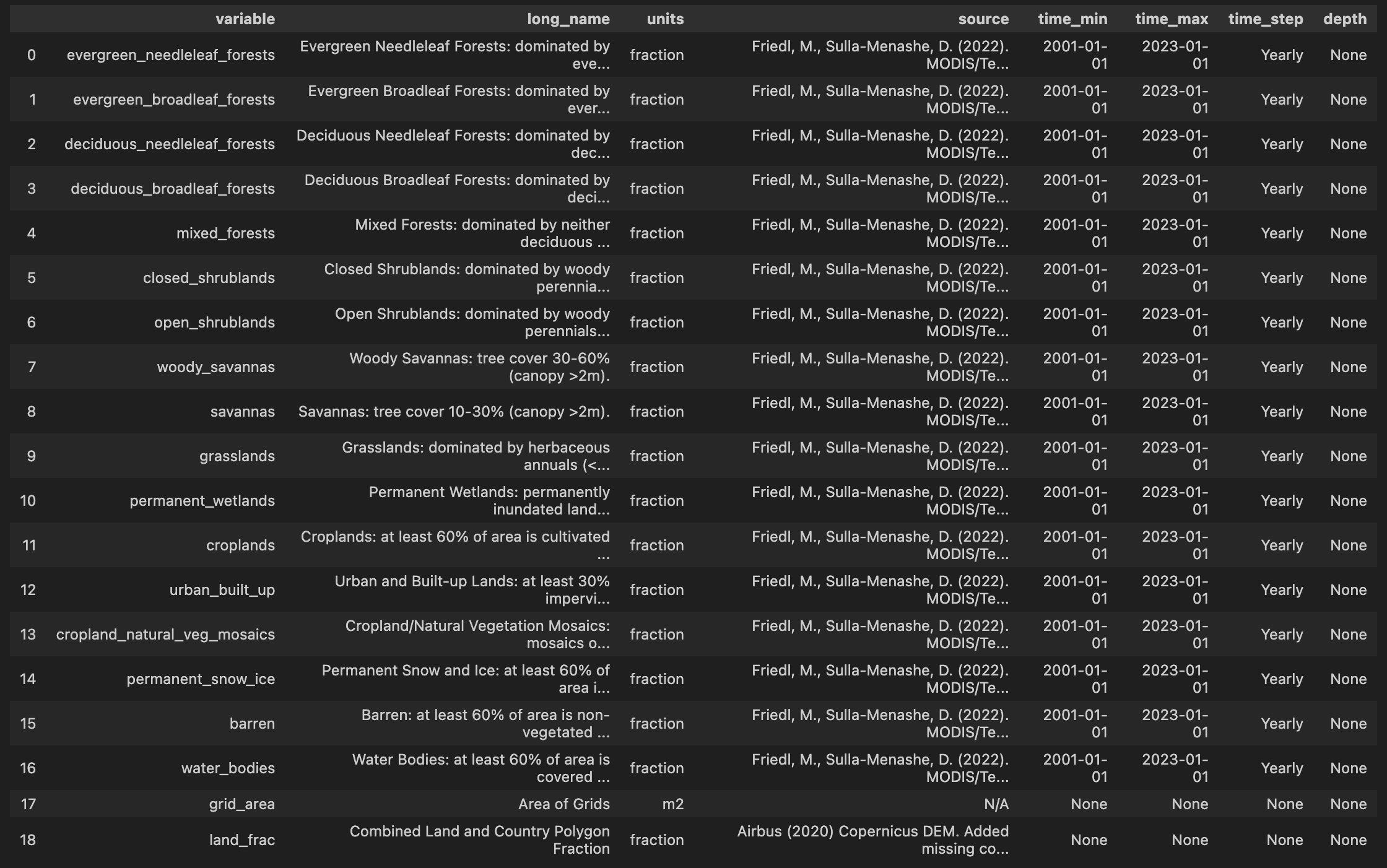
- View dataset metadata
ssm.list_variables("atlas/B.land.cover.2001-2023.a.nc")
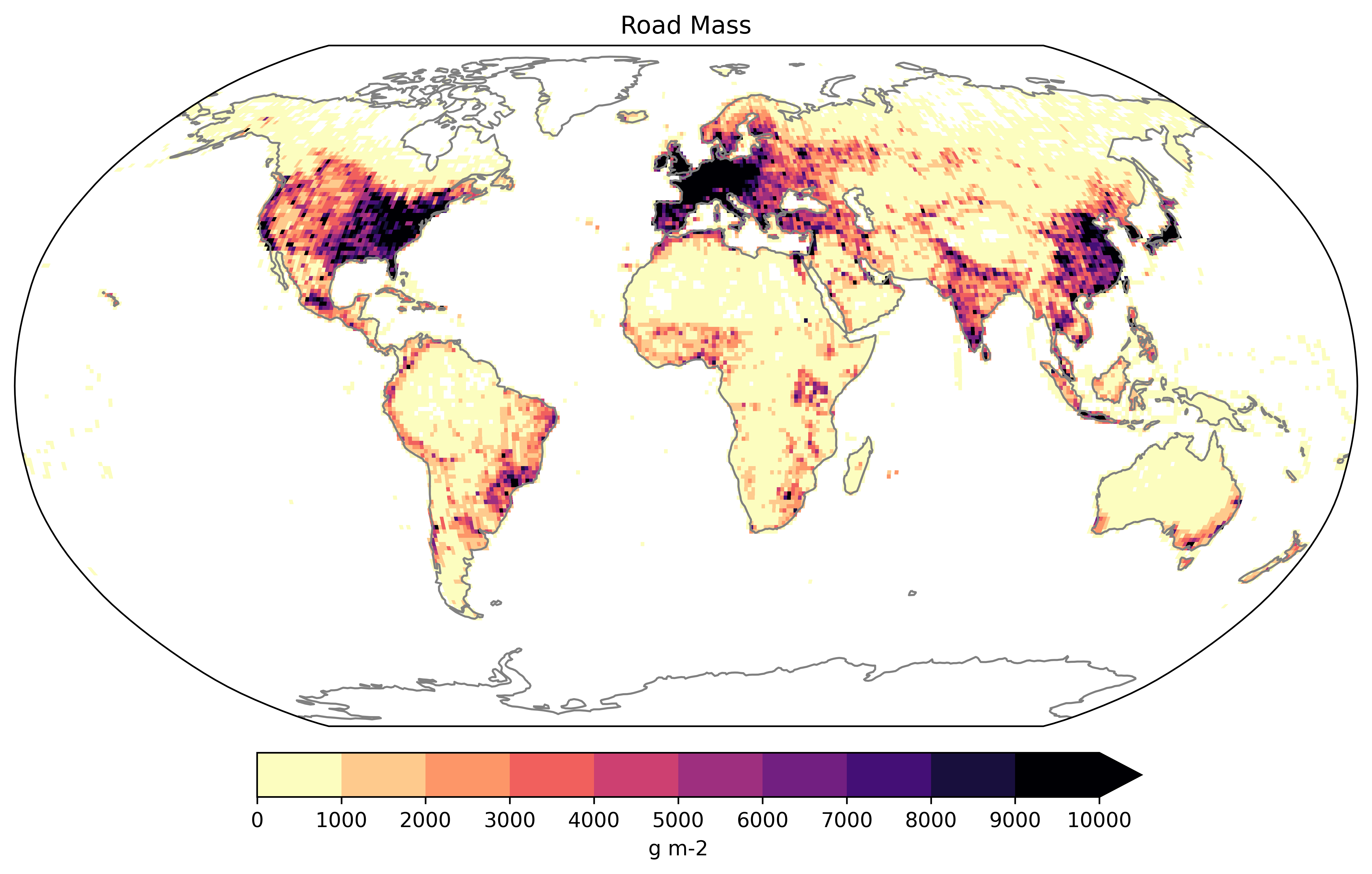
- Visualize data on the map
# Load data
netcdf_file = "atlas/T.transportation.roads.nc"
ssm.plot_map(dataset=netcdf_file,variable="roads_gross", color='magma_r', title='Gross Road Mass', label='g m-2', vmin=0, vmax=1e4, extend_max=True)
- Quick mathematical operation
# Load data
netcdf_file = "atlas/T.transportation.roads.nc"
# Perform the operation
ssm.divide_variables(dataset=netcdf_file, variable1="road_length", variable2="grid_area", new_variable_name="road_density")
Ready to get started? Dive into the function docs below or read The SESAME Human-Earth Atlas paper for inspiration!
Converts point data from a shapefile or GeoDataFrame into a gridded netCDF dataset.
Parameters
- point_data : GeoDataFrame or str. Input point data to be gridded. Can be either a GeoDataFrame or a path to a point shapefile (.shp).
- variable_name : str, optional. Name of the variable to include in the netCDF attributes metadata. Defaults to:
- The unique entries in the
attr_fieldcolumn if specified. - The input filename without extension if
attr_fieldandvariable_nameare not specified.
- The unique entries in the
- long_name : str, optional. A descriptive name for the variable, added to the netCDF metadata. Behaves the same as
variable_nameifattr_fieldis specified. Defaults to the input filename without extension if unspecified. - units : str, optional. Units of the data variable to include in the netCDF metadata. Default is "value/grid-cell".
- source : str, optional. String describing the original source of the input data. This will be added to the netCDF metadata.
- time : str, optional. Time dimension for the output netCDF. If specified, the output will include a time dimension with the value provided. Default is None (spatial, 2D netCDF output).
- resolution : float, optional. Desired resolution for the grid cells in the output dataset. Default is 1 degree.
- agg_column : str, optional. Column name in the shapefile or GeoDataFrame specifying the values to aggregate in each grid cell. Defaults to counting the number of points per grid cell.
- agg_function : str, optional. Aggregation method for combining values in each grid cell. Options include:
- 'sum' (default): Sums all point values.
- 'max': Takes the maximum value.
- 'min': Takes the minimum value.
- 'std': Computes the standard deviation.
- attr_field : str, optional. Column name in the shapefile or GeoDataFrame specifying the variable names for multiple data types.
- output_directory : str, optional. Directory where the output NetCDF file will be saved. If None, but output_filename is True, the file will be saved in the current working directory.
- output_filename : str, optional. Name of the output NetCDF file (without the
.ncextension). If not provided:- Uses the input shapefile name if a shapefile path is given.
- Saves as
"gridded_points.nc"if a GeoDataFrame is provided as input.
- normalize_by_area : bool, optional. If True, normalizes the grid values by area (e.g., converts to value per square meter). Default is False.
- zero_is_value : bool, optional. If True, treats zero values as valid data rather than as no-data. Default is False.
- verbose : bool, optional. If True, prints information about the process, such as global sum of values before and after gridding. Default is False.
Returns
- xarray.Dataset. Transformed dataset with gridded data derived from the input point data.
Notes
- The function supports input in the form of a shapefile or GeoDataFrame containing point data.
- If points lie exactly on a grid boundary, they are shifted by 0.0001 degrees in both latitude and longitude to ensure assignment to a grid cell.
- The function creates a netCDF file, where data variables are aggregated based on the
agg_columnandagg_function.
Example
>>> point_2_grid(point_data=shapefile_path,
... variable_name="airplanes",
... long_name="Airplanes Count",
... units="airport/grid-cell",
... source="CIA",
... resolution=1,
... verbose=True
... )
Converts line data from a shapefile or GeoDataFrame into a gridded netCDF dataset.
Parameters
- line_data : GeoDataFrame or str. Input lines data to be gridded. Can be either a GeoDataFrame or a path to a line/polyline shapefile (.shp).
- variable_name : str, optional. Name of the variable to include in the netCDF attributes metadata. Defaults to:
- The unique entries in the
attr_fieldcolumn if specified. - The input filename without extension if
attr_fieldandvariable_nameare not specified.
- The unique entries in the
- long_name : str, optional. A descriptive name for the variable, added to the netCDF metadata. Behaves the same as
variable_nameifattr_fieldis specified. Defaults to the input filename without extension if unspecified. - units : str, optional. Units of the data variable to include in the netCDF metadata. Default is "meter/grid-cell".
- source : str, optional. String describing the original source of the input data. This will be added to the netCDF metadata.
- time : str, optional. Time dimension for the output netCDF. If specified, the output will include a time dimension with the value provided. Default is None (spatial, 2D netCDF output).
- resolution : float, optional. Desired resolution for the grid cells in the output dataset. Default is 1 degree.
- agg_column : str, optional. Column name in the shapefile or GeoDataFrame specifying the values to aggregate in each grid cell. Defaults to summing the lengths of intersected lines per grid cell.
- agg_function : str, optional. Aggregation method for combining values in each grid cell. Options include:
- 'sum' (default): Sums all line values.
- 'max': Takes the maximum value.
- 'min': Takes the minimum value.
- 'std': Computes the standard deviation.
- attr_field : str, optional. Column name in the shapefile or GeoDataFrame specifying the variable names for multiple data types.
- output_directory : str, optional. Directory where the output NetCDF file will be saved. If None, but output_filename is True, the file will be saved in the current working directory.
- output_filename : str, optional. Name of the output NetCDF file (without the
.ncextension). If not provided:- Uses the input shapefile name if a shapefile path is given.
- Saves as
"gridded_lines.nc"if a GeoDataFrame is provided as input.
- normalize_by_area : bool, optional. If True, normalizes the variable in each grid cell by the area of the grid cell (e.g., converts to value per square meter). Default is False.
- zero_is_value : bool, optional. If True, treats zero values as valid data rather than as no-data. Default is False. If True, treats zero values as valid data rather than as no-data. Default is False.
- verbose : bool, optional. If True, prints information about the process, such as global sum of values before and after gridding. Default is False.
Returns
- xarray.Dataset. Transformed dataset with gridded data derived from the input line data.
Notes
- The function supports input in the form of a shapefile or GeoDataFrame containing line data.
- Line lengths are calculated and aggregated based on the specified
agg_columnandagg_function. - If lines intersect a grid boundary, their contributions are divided proportionally among the intersected grid cells.
- The function creates a netCDF file, where data variables are aggregated and stored with metadata.
Example
>>> line_2_grid(line_data=shapefile_path,
... variable_name="roads",
... long_name="Roads Length",
... units="meter/grid-cell",
... source="OpenStreetMap",
... resolution=1,
... agg_function="sum",
... verbose=True)
... )
Converts polygon data from a shapefile or GeoDataFrame into a gridded netCDF dataset.
Parameters
- polygon_data : GeoDataFrame or str. Input polygons data to be gridded. Can be either a GeoDataFrame or a path to a polygons shapefile (.shp).
- variable_name : str, optional. Name of the variable to include in the netCDF attributes metadata. Defaults to:
- The unique entries in the
attr_fieldcolumn if specified. - The input filename without extension if
attr_fieldandvariable_nameare not specified.
- The unique entries in the
- long_name : str, optional. A descriptive name for the variable, added to the netCDF metadata. Behaves the same as
variable_nameifattr_fieldis specified. Defaults to the input filename without extension if unspecified. - units : str, optional. Units of the data variable to include in the netCDF metadata. Default is "m2/grid-cell".
- source : str, optional. String describing the original source of the input data. This will be added to the netCDF metadata.
- time : str, optional. Time dimension for the output netCDF. If specified, the output will include a time dimension with the value provided. Default is None (spatial, 2D netCDF output).
- resolution : float, optional. Desired resolution for the grid cells in the output dataset. Default is 1 degree.
- attr_field : str, optional. Column name in the shapefile or GeoDataFrame specifying the variable names for multiple data types.
- fraction : bool, optional. If True, calculates the fraction of each polygon within each grid cell. The output values will range from 0 to 1. Default is False.
- agg_function : str, optional. Aggregation method for combining values in each grid cell. Default is 'sum'. Options include:
- 'sum': Sum of values.
- 'max': Maximum value.
- 'min': Minimum value.
- 'std': Standard deviation.
- output_directory : str, optional. Directory where the output NetCDF file will be saved. If None, but output_filename is True, the file will be saved in the current working directory.
- output_filename : str, optional. Name of the output NetCDF file (without the
.ncextension). If not provided:- Uses the input shapefile name if a shapefile path is given.
- Saves as
"gridded_polygons.nc"if a GeoDataFrame is provided as input.
- normalize_by_area : bool, optional. If True, normalizes the grid values by area (e.g., converts to value per square meter). Default is False.
- zero_is_value : bool, optional. If True, treats zero values as valid data rather than as no-data. Default is False.
- verbose : bool, optional. If True, prints information about the process, such as global sum of values before and after gridding. Default is False.
Returns
- xarray.Dataset. Transformed dataset with gridded data derived from the input polygon data.
Notes
- The function supports input in the form of a shapefile or GeoDataFrame containing polygon data.
- Polygon areas are calculated and aggregated based on the specified
attr_fieldandagg_function. - If the
fractionparameter is True, the fraction of each polygon in each grid cell will be computed, with values ranging from 0 to 1. - The function creates a netCDF file, where data variables are aggregated and stored with metadata.
Example
>>> poly_2_grid(polygon_data=shapefile_path,
... units="fraction",
... source="The new global lithological map database GLiM",
... resolution=1,
... attr_field="Short_Name",
... fraction="yes",
... verbose=True
... )
Converts raster data (TIFF or netCDF) into a re-gridded xarray dataset.
Parameters
- raster_data : str. Path to the input raster data file. This can be a string path to a TIFF (.tif) file, a string path to a NetCDF (.nc or .nc4) file or An already loaded xarray.Dataset object.
- If
raster_datais a NetCDF file or an xarray.Dataset, thenetcdf_variableparameter must also be provided to specify which variable to extract.
- If
- agg_function : str. Aggregation method to apply when re-gridding. Supported values are 'SUM', 'MEAN', or 'MAX'.
- variable_name : str. Name of the variable to include in the output dataset.
- long_name : str. Descriptive name for the variable.
- units : str, optional. Units for the variable. Default is "value/grid-cell".
- source : str, optional. Source information for the dataset. Default is None.
- time : str or None, optional. Time stamp or identifier for the data. Default is None.
- resolution : int or float, optional. Desired resolution of the grid cells in degree in the output dataset. Default is 1.
- netcdf_variable : str, optional. Name of the variable to extract from the netCDF file, if applicable. Required for netCDF inputs.
- output_directory : str, optional. Directory where the output NetCDF file will be saved. If None, but output_filename is True, the file will be saved in the current working directory.
- output_filename : str, optional. Name of the output NetCDF file (without the
.ncextension). If not provided:- Uses
variable_nameif it is specified. - Defaults to
regridded.ncif none of the above are provided.
- Uses
- padding : str, optional. Padding strategy ('symmetric' or 'end').
- zero_is_value : bool, optional. Whether to treat zero values as valid data rather than as no-data. Default is False.
- normalize_by_area : bool, optional. Whether to normalize grid values by area (e.g., convert to value per square meter). Default is False.
- verbose : bool, optional. If True, prints the global sum of values before and after re-gridding. Default is False.
Returns
- xarray.Dataset. Re-gridded xarray dataset containing the processed raster data.
Notes
This function supports raster data in TIFF or netCDF format and performs re-gridding based on
the specified agg_function. The output dataset will include metadata such as the variable name,
long name, units, and optional source and time information.
Example
>>> grid_2_grid(raster_path=pop_path,
... agg_function="sum",
... variable_name="population_count",
... long_name="Total Population",
... units="people per grid",
... source="WorldPop",
... resolution=1,
... time="2020-01-01",
... verbose="yes"
... )
Convert tabular data to a gridded dataset by spatially distributing values based on a NetCDF variable and a tabular column.
Parameters:
- surrogate_data : xarray.Dataset or str. xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded
into an xarray.Dataset. The dataset must include the variable specified in
surrogate_variable. - surrogate_variable : str. Variable name in the NetCDF or xarray dataset used for spatial distribution.
- tabular_data : pandas.DataFrame or str. Tabular dataset as a pandas DataFrame or a path to a CSV file. If a file path is provided, it will be
automatically loaded into a DataFrame. The data must include a column named "ISO3" representing country codes.
If not present, use the
add_iso3_columnutility function to convert country names to ISO3 codes. - tabular_column : str. Column name in the tabular dataset with values to be spatially distributed.
- variable_name : str, optional. Name of the variable. Default is None.
- long_name : str, optional. A long name for the variable. Default is None.
- units : str, optional. Units of the variable. Default is 'value/grid'.
- source : str, optional. Source information, if available. Default is None.
- time : str, optional. Time information for the dataset.
- output_directory : str, optional. Directory where the output NetCDF file will be saved. If None, but output_filename is True, the file will be saved in the current working directory.
- output_filename : str, optional. Name of the output NetCDF file (without the
.ncextension). If not provided:- Uses
variable_nameif it is specified. - Falls back to
long_nameortabular_columnifvariable_nameis not given. - Defaults to
gridded_table.ncif none of the above are provided.
- Uses
- zero_is_value: bool, optional. If the value is True, then the function will treat zero as an existent value and 0 values will be considered while calculating mean and STD.
- normalize_by_area : bool, optional. Whether to normalize grid values by area (e.g., convert to value per square meter). Default is False.
- eez : bool, optional. If set to True, the function converts the jurisdictional Exclusive Economic Zone (EEZ) values to a spatial grid.
- verbose: bool, optional. If True, the global gridded sum of before and after re-gridding operation will be printed. If any jurisdiction where surrogate variable is missing and tabular data is evenly distributed over the jurisdiction, the ISO3 codes of evenly distributed countries will also be printed.
Returns:
- xarray.Dataset. Resulting gridded dataset after spatial distribution of tabular values.
Example
>>> table_2_grid(surrogate_data=netcdf_file_path,
... surrogate_variable="railway_length",
... tabular_data=csv_file_path,
... tabular_column="steel",
... variable_name="railtract_steel",
... long_name="'Railtrack Steel Mass'",
... units="g m-2",
... source="Matitia (2022)",
... normalize_by_area="yes",
... verbose="yes"
... )
Process gridded data from an xarray Dataset to generate tabular data for different jurisdictions.
Parameters:
- grid_data : xarray.Dataset or str. xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variables : str, optional. Variables name to be processed. It can either be one variable or list of variables. If None, all variables in the dataset (excluding predefined ones) will be considered.
- time : str, optional. Time slice for data processing. If provided, the nearest time slice is selected. If None, a default time slice is used.
- resolution : float, optional. Resolution of gridded data in degree. Default is 1 degree.
- grid_area : str, optional. Indicator to consider grid area during processing. If 'YES', the variable is multiplied by grid area.
- aggregation : str, optional. Aggregation level for tabular data. If 'continent', the data will be aggregated at the continent level.
- agg_function : str, optional, default 'sum'. Aggregation method. Options: 'sum', 'mean', 'max', 'min', 'std'.
- verbose : bool, optional. If True, the function will print the global sum of values before and after aggregation.
Returns:
df : pandas DataFrame. Tabular data for different jurisdictions, including ISO3 codes, variable values, and optional 'Year' column.
Convert country names in a DataFrame column to their corresponding ISO3 country codes.
This function reads a JSON file containing country names and their corresponding ISO3 codes, then maps the values from the specified column in the DataFrame to their ISO3 codes based on the JSON data. The resulting ISO3 codes are added as a new column named 'ISO3'.
Parameters
- df (pandas.DataFrame): The DataFrame containing a column with country names.
- column (str): The name of the column in the DataFrame that contains country names.
Returns
- pandas.DataFrame: The original DataFrame with an additional 'ISO3' column containing the ISO3 country codes.
Raises:
- FileNotFoundError: If the JSON file containing country mappings cannot be found.
- KeyError: If the specified column is not present in the DataFrame.
Example
>>> add_iso3_column(df=dataframe,
... column="Country"
... )
Create a histogram for an array variable in an xarray dataset. Optionally remove outliers and apply log transformations.
Parameters:
- dataset : xarray.Dataset or str, xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variable: str, the name of the variable to plot.
- time: str, optional, the time slice to plot.
- bin_size: int, optional, the number of bins in the histogram.
- color: str, optional, the color of the histogram bars.
- plot_title: str, optional, the title for the plot.
- x_label: str, optional, the label for the x-axis.
- remove_outliers: bool, optional, whether to remove outliers.
- log_transform: str, optional, the type of log transformation ('log10', 'log', 'log2').
- output_dir : str, optional, Directory path to save the output figure. If not provided, the figure is saved in the current working directory.
- filename : str, optional, Filename (with extension) for saving the figure. If not provided, the plot is saved as "output_histogram.png".
Returns:
- None, displays the plot and optionally saves it to a file.
Example
>>> plot_histogram(dataset=dataset,
... variable="railway_length",
... bin_size=30,
... color='blue',
... plot_title="Histogram of Railway Length"
... )
Create a scatter plot for two variables in an xarray dataset. Optionally remove outliers and apply log transformations.
Parameters:
- variable1 : str, name of the variable to be plotted on the x-axis. Must be present in
dataset. - variable2 : str, name of the variable to be plotted on the y-axis. If
dataset2is provided, this variable will be extracted fromdataset2; otherwise, it must exist indataset. - dataset : xarray.Dataset or str, the primary dataset or a path to a NetCDF file. This dataset must contain the variable specified by
variable1, which will be used for the x-axis. - dataset2 : xarray.Dataset or str, optional, a second dataset or a path to a NetCDF file containing the variable specified by
variable2(for the y-axis). If not provided,datasetwill be used for both variables. - time: str, optional, the time slice to plot.
- color: str, optional, the color map of the scatter plot.
- x_label: str, optional, the label for the x-axis.
- y_label: str, optional, the label for the y-axis.
- plot_title: str, optional, the title for the plot.
- remove_outliers: bool, optional, whether to remove outliers from the data.
- log_transform_1: str, optional, the type of log transformation for variable1 ('log10', 'log', 'log2').
- log_transform_2: str, optional, the type of log transformation for variable2 ('log10', 'log', 'log2').
- equation : bool, optional, ff True, fits and displays a linear regression equation.
- output_dir : str, optional, Directory path to save the output figure. If not provided, the figure is saved in the current working directory.
- filename : str, optional, Filename (with extension) for saving the figure. If not provided, the plot is saved as "output_scatter.png".
Returns:
- None, displays the plot and optionally saves it to a file.
Example
>>> plot_scatter(dataset=ds_road,
... variable1="roads_gross",
... variable2="buildings_gross",
... dataset2=ds_build,
... color='blue',
... plot_title="Building vs Road",
... remove_outliers=True,
... log_transform_1="log10",
... log_transform_2="log10"
... )
Create a line plot and/or area plot for a time series data variable.
Parameters:
- dataset : xarray.Dataset or str, xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variable: str, the name of the variable to plot.
- agg_function: str, the operation to apply ('sum', 'mean', 'max', 'std').
- plot_type: str, optional, the type of plot ('line', 'area', 'both'). Default is 'both'.
- color: str, optional, the color of the plot. Default is 'blue'.
- plot_label: str, optional, the label for the plot. Default is 'Area Plot'.
- x_label: str, optional, the label for the x-axis. Default is 'Year'.
- y_label: str, optional, the label for the y-axis. Default is 'Value'.
- plot_title: str, optional, the title of the plot. Default is 'Time Series Plot'.
- smoothing_window: int, optional, the window size for rolling mean smoothing.
- output_dir : str, optional, Directory path to save the output figure. If not provided, the figure is saved in the current working directory.
- filename : str, optional, Filename (with extension) for saving the figure. If not provided, the plot is saved as "output_time_series.png".
Returns:
- None, displays the plot and optionally saves it to a file.
Example
>>> plot_time_series(variable="buildings_gross",
... dataset=ds_build,
... agg_function='sum',
... plot_type='both',
... color='blue',
... x_label='Year',
... y_label='Value',
... plot_title='Time Series Plot'
... )
Create a hexbin plot for two variables in an xarray dataset.
Parameters:
- dataset : xarray.Dataset or str, the primary dataset or a path to a NetCDF file. This dataset must contain the variable specified by
variable1, which will be used for the x-axis. - variable1 : str, name of the variable to be plotted on the x-axis. Must be present in
dataset. - variable2 : str, name of the variable to be plotted on the y-axis. If
dataset2is provided, this variable will be extracted fromdataset2; otherwise, it must exist indataset. - dataset2 : xarray.Dataset or str, optional, a second dataset or a path to a NetCDF file containing the variable specified by
variable2(for the y-axis). If not provided,datasetwill be used for both variables. - time: str, optional, the time slice to plot.
- color: str, optional, the color map of the hexbin plot.
- grid_size: int, optional, the number of hexagons in the x-direction.
- x_label: str, optional, the label for the x-axis.
- y_label: str, optional, the label for the y-axis.
- plot_title: str, optional, the title for the plot.
- remove_outliers: bool, optional, whether to remove outliers from the data.
- log_transform_1: str, optional, the type of log transformation for variable1 ('log10', 'log', 'log2').
- log_transform_2: str, optional, the type of log transformation for variable2 ('log10', 'log', 'log2').
- output_dir : str, optional, Directory path to save the output figure. If not provided, the figure is saved in the current working directory.
- filename : str, optional, Filename (with extension) for saving the figure. If not provided, the plot is saved as "output_hexbin.png".
Returns:
- None, displays the map and optionally saves it to a file.
Example
>>> plot_hexbin(dataset=ds_road,
... variable1="roads_gross",
... variable2="buildings_gross",
... dataset2=ds_build,
... color='blue',
... plot_title="Building vs Road"
... )
Plots a 2D map of a variable from an xarray Dataset or NetCDF file with customizable colorbar, projection, and map appearance.
Parameters
- dataset : xarray.Dataset. or str. xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variable : str. Name of the variable in the xarray Dataset to plot.
- color : str, default 'hot_r'. Matplotlib colormap name for the plot (discrete color scale).
- title : str, default ''. Title of the map.
- label : str, default ''. Label for the colorbar.
- time: str, optional, the time slice to plot.
- vmin : float, optional. Minimum data value for the colorbar range. If not provided, the minimum of the variable is used.
- vmax : float, optional. Maximum data value for the colorbar range. If not provided, the maximum of the variable is used.
- extend_min : bool, default False. If True, includes values below
vminin the first color class and shows a left arrow on the colorbar. - extend_max : bool, default False. If True, includes values above
vmaxin the last color class and shows a right arrow on the colorbar. - levels : int or list of float, default 10. Either the number of color intervals or a list of explicit interval boundaries.
- out_bound : bool, default True. Whether to display the outer boundary (spine) of the map projection.
- remove_ata : bool, default False. If True, removes Antarctica from the map by excluding data below 60°S latitude.
- output_dir : str, optional. Directory path to save the output figure. If not provided, the figure is saved in the current working directory.
- filename : str, optional. Filename (with extension) for saving the figure. If not provided, the plot is saved as "output_plot.png".
- show : bool, True. Whether or not show the map
Notes
- If both
extend_minandextend_maxare False, the dataset is clipped strictly within [vmin, vmax]. - The colorbar will use arrows to indicate out-of-bound values only if
extend_minorextend_maxis True. - Tick formatting on the colorbar is:
- Two decimal places if (vmax - vmin) <= 10.
- If
remove_atais True, the colorbar is placed slightly higher to avoid overlap with the map.
Returns:
- Axes class of the map, optionally displays the map and saves it to a file.
Example
>>> plot_map(
... dataset=ds.isel(time=-1),
... variable='npp',
... vmin=0,
... vmax=1200,
... extend_max=True,
... color='Greens',
... levels=10,
... remove_ata=True,
... title='Net Primary Productivity',
... label='gC/m²/year',
... filename='npp_map.png'
... )
Plots a choropleth map of countries using a specified data column and a world shapefile.
Parameters:
- tabular_data : pandas.DataFrame or str. Input table containing country-level data. Can be either:
- A pandas DataFrame with the required
column - A string path to a CSV file, which will be automatically read into a DataFrame
- A pandas DataFrame with the required
- column : str. Name of the column in the dataframe to visualize.
- title : str, optional. Title of the map. Default is an empty string.
- label : str, optional. Label for the colorbar. Default is an empty string.
- color : str, optional. Name of the matplotlib colormap to use. Default is 'viridis'.
- levels : int or list of float, optional. Number of color levels (if int) or list of bin edges (if list). Default is 10.
- remove_ata : bool, optional. Whether to remove Antarctica ('ATA') from the data. Default is False.
- out_bound : bool, optional. Whether to display map boundaries (spines). Default is True.
- vmin : float or None, optional. Minimum value for the colormap. If None, calculated from the data.
- vmax : float or None, optional. Maximum value for the colormap. If None, calculated from the data.
- extend_min : bool, optional. Whether to extend the colorbar below
vmin. Default is False. - extend_max : bool, optional. Whether to extend the colorbar above
vmax. Default is False. - output_dir : str, optional. Directory path to save the output figure. If not provided, the figure is saved in the current working directory.
- filename : str, optional. Filename (with extension) for saving the figure. If not provided, the plot is saved as "output_country_plot.png".
Returns:
- None, displays the map and optionally saves it to a file.
Example
>>> plot_country(tabular_data="country_data.csv",
... column="population",
... title="Population of Countries",
... label="Population",
... color='viridis'
... )
Sum specified variables in the xarray dataset. If no variables are specified, sum all variables except those starting with 'grid_area'. Fill NaNs with zero before summing, and convert resulting zeros back to NaNs.
Parameters:
- dataset: xarray.Dataset. or str, xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variables: list of str, the names of the variables to sum. If None, sum all variables except those starting with 'grid_area' and 'land_frac'.
- new_variable_name: str, optional, the name of the new variable to store the sum.
- time: optional, a specific time slice to select from the dataset.
Returns:
- xarray.Dataset. with the summed variable.
Example
>>> sum_variables(dataset=ds,
... variables=["roads_gross", "buildings_gross"],
... new_variable_name="gross_mass"
... )
Subtract one variable from another in the xarray dataset. Fill NaNs with zero before subtracting, and convert resulting zeros back to NaNs.
Parameters:
- dataset: xarray.Dataset. or str, xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variable1: str, the name of the variable to subtract from.
- variable2: str, the name of the variable to subtract.
- new_variable_name: str, optional, the name of the new variable to store the result.
- time: optional, a specific time slice to select from the dataset.
Returns:
- xarray.Dataset. with the resulting variable.
Example
>>> subtract_variables(dataset=ds,
... variable1="precipitation",
... variable2="evaporation",
... new_variable_name="net_water_gain"
... )
Divide one variable by another in the xarray dataset. Fill NaNs with zero before dividing, and convert resulting zeros back to NaNs.
Parameters:
- dataset: xarray.Dataset. or str, xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variable1: str, the name of the variable to be divided (numerator).
- variable2: str, the name of the variable to divide by (denominator).
- new_variable_name: str, optional, the name of the new variable to store the result.
- time: optional, a specific time slice to select from the dataset.
Returns:
- xarray.Dataset. with the resulting variable.
Example
>>> divide_variables(dataset=ds,
... variable1="road_length",
... variable2="grid_area",
... new_variable_name="road_density"
... )
Multiply specified variables in the xarray dataset. If no variables are specified, multiply all variables. Fill NaNs with one before multiplying, and convert resulting ones back to NaNs.
Parameters:
- dataset: xarray.Dataset. or str, xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variables: list of str, the names of the variables to multiply. If None, multiply all variables, excluding the "grid_area" and "land_frac" variables included in the dataset.
- new_variable_name: str, optional, the name of the new variable to store the product.
- time: optional, a specific time slice to select from the dataset.
Returns:
- xarray.Dataset. with the resulting variable.
Example
>>> multiply_variables(
... dataset=ds,
... variables=["crop_area", "yield_per_hectare"],
... new_variable_name="total_crop_yield"
... )
Average specified variables in the xarray dataset. If no variables are specified, average all variables except those starting with 'grid_area'. Fill NaNs with zero before averaging, and convert resulting zeros back to NaNs.
Parameters:
- dataset: xarray.Dataset. or str, xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variables: list of str, the names of the variables to average. If None, average all variables except those starting with 'grid_area' and 'land_frac'.
- new_variable_name: str, optional, the name of the new variable to store the average.
- time: optional, a specific time slice to select from the dataset.
Returns:
- xarray.Dataset. with the averaged variable.
Example
>>> average_variables(dataset=ds,
... variables=["roads_gross", "buildings_gross"],
... new_variable_name="average_gross"
... )
Extract information about variables and dimensions from a NetCDF dataset.
Parameters
- netcdf_file : xarray.Dataset or str. xarray dataset or a path to a NetCDF file. If a file path is provided, it will be automatically loaded into an xarray.Dataset.
- variable_name : str, optional. The prefix or complete name of the variable to filter. If not provided, all variables are included.
Returns
- tuple, A tuple containing lists of dimensions, short names, long names, units, & time values (if 'time' exists).
Example
>>> get_netcdf_info(netcdf_file=netcdf_file_path,
... variable_name="railway_length"
... )
List all NetCDF files in a directory and count the number of variables in each.
Parameters
directory : str. Path to the directory containing NetCDF files.
Returns
pd.DataFrame. A DataFrame with file names and the number of variables in each file.
Example
>>> atlas(directory)
Extract metadata for each variable in a NetCDF dataset.
Parameters
- data : str, os.PathLike, or xarray.Dataset. Path to a NetCDF file or an xarray.Dataset object.
Returns
- pd.DataFrame. A DataFrame containing variable names, long names, units, sources, time range (start and end), time resolution (step), and depth values (if present as a variable).
Example
>>> info(netcdf_path)